Blind demixing methods for recovering dense neuronal morphology from barcode imaging data
Abstract
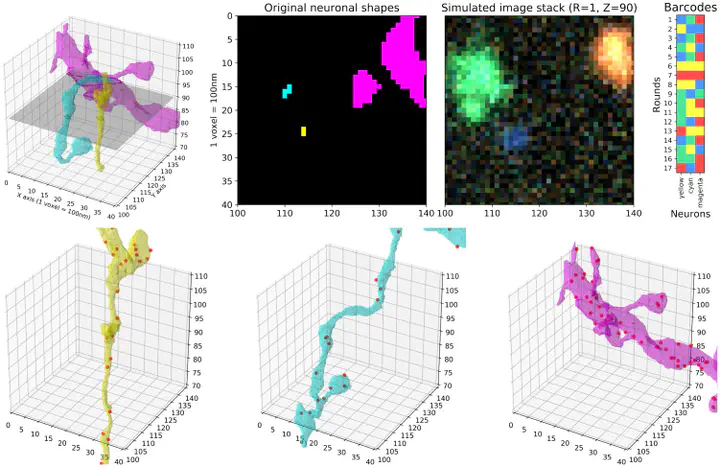
Cellular barcoding methods offer the exciting possibility of ‘infinite-pseudocolor’ anatomical reconstruction—i.e., assigning each neuron its own random unique barcoded ‘pseudocolor,’ and then using these pseudocolors to trace the microanatomy of each neuron. Here we use simulations, based on densely-reconstructed electron microscopy microanatomy, with signal structure matched to real barcoding data, to quantify the feasibility of this procedure. We develop a new blind demixing approach to recover the barcodes that label each neuron, and validate this method on real data with known barcodes. We also develop a neural network which uses the recovered barcodes to reconstruct the neuronal morphology from the observed fluorescence imaging data, ‘connecting the dots’ between discontiguous barcode amplicon signals. We find that accurate recovery should be feasible, provided that the barcode signal density is sufficiently high. This study suggests the possibility of mapping the morphology and projection pattern of many individual neurons simultaneously, at high resolution and at large scale, via conventional light microscopy., In situ barcode sequencing allows us to simultaneously locate many neurons in intact brain tissues, albeit at modest spatial resolution. By increasing the barcode density, high-resolution neuronal morphology reconstruction from such data might be possible. Here we use simulations to study this possibility, while addressing the computational challenges in analyzing such data. We developed a novel blind demixing method that uses fluorescent images and identifies the unknown barcodes used to label the neurons with high accuracy. Further, we developed a neural network which can reconstruct the morphology for these labeled neurons from the observed ‘pointilistic’ imaging data. We show that under both high- and low-resolution optical settings, our methods can successfully extract the morphologies for many labeled neurons. The results from this theoretical study suggest that it may be feasible to map the morphology and projection pattern of many individual neurons simultaneously, at high resolution and at large scale, via conventional light microscopy.